
TLDR
We need to import several CSV or TXT files and merge them into one data frame in R. Regardless of what function we use to import the files, vectorising the operation using
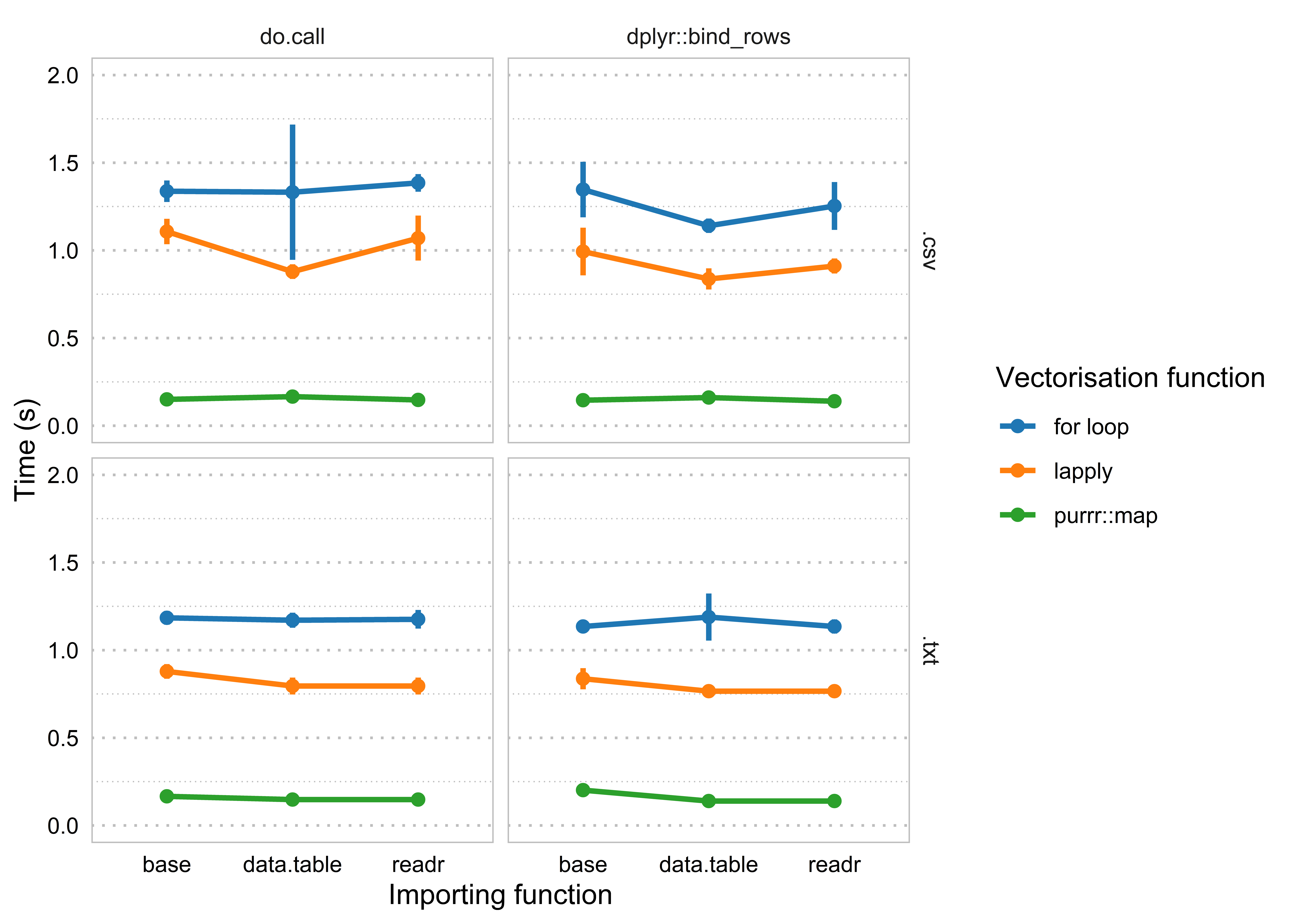
purrr::mapin combination withdo.callordplyr::bind_rowsis the most time-efficient method (~25 ms importing 50 files with 10,000 rows each), compared to for loops (~220ms) or usinglapply(~123 ms).data.table::freadis the fastest function for importing data. Importing TXT files is slightly faster than importing CSV files.
Why this post
To analyse data in any programming environment, one must first import some data. Sometimes, the data we want to analyse are distributed across several files in the same folder. I work with eye-tracking data from toddlers. This means that I work with multiple files that have many rows. At 120 Hz sampling frequency, we take ~8.33 samples per second. A session for one participants can take up to 10 minutes. So these files are somewhat big. These data also tend to be messy, requiring a lot of preprocessing. This means that I need to import the same large files many times during the same R session when wrangling my way through the data, which takes a few seconds. After some iterations, it can be annoying. I have decided to invest all my lost time into analysing what method for importing and merging large files is the fastest in R so that the universe and I are even again.
Below I provide several options for importing data from the different files, using base R and tidyverse, among other tools. I will compare how long it takes to import and merge data using each method under different circumstances. You can find the whole code here in case you want to take a look, reproduce it or play with it1.
1 Ironically, this code is super inefficient and messy. It takes ages to run, and has been written by copy-pasting multiple times. I didn’t feel like doing anything more elegant. Also, I don’t know how. Help yourself.
How can I import large files and merge them?
So we have some files in a folder. All files have the same number of columns, the same column names, and are in the same format. I assume that data are tabular (i.e., in the shape of a rectangle defined by rows and columns). I also assume that data are stored as Comma-Separated Values (.csv) or Tab-separated Text (.txt or .tsv), as these formats are the most reproducible.
We to import all files and bind their rows together to form a unique long data frame. There are multiple combinations of functions we can use. Each function comes with a different package and does the job in different ways. Next, I will show some suggestions, but first let’s create some data. We are creating 50 datasets with 10 columns and 10,000 rows in .txt format. The variables included are numeric and made of 0s and 1s. There is also a column that identifies the data set. These files are created in a temporary directory using the temp.dir function for reproducibility. After closing you R session, this directory and all of its contents will disappear.
Base R: for loops
for loops are one of the fundamental skills in many programming languages. The idea behind for loops is quite intuitive: take a vector or list of length n, and apply a series of functions to each element in order. First, to element 1 and then to element 2, and so on, until we get to element n. Then, the loop ends. We will first make a vector with the paths of our files, and then apply the read.delim function to each element of the vector (i.e., to each path). Every time we import a file, we store the resulting data frame as an element of a list. After the loop finishes, we merge the rows of all element of the list using a combination of the functions do.call and rbind.
Base R: lapply
We will use the functions read.delim and read.csv in combination with the function lapply. The former are well known. The later is part of a family of functions (together with sapply, mapply, and some others I can’t remember) that take two arguments: a list and a function, which will be applied over each element of the list in parallel (i.e., in a vectorised way).
Tidyverse
The tidyverse is a family of packages that suggests a workflow when working in R. The use of pipes (%>%) is one of its signature moves, which allow you to chain several operations applied on the same object within the same block of code. In contrast, base R makes you choose between applying several functions to the same object in different blocks of code, or applying those functions in a nested way, so that the first functions you read are those applied the last to your object (e.g., do.call(rbind, as.list(data.frame(x = "this is annoying", y = 1:100)))). We will use a combination of the dplyr and purrr packages to import the files listed in a vector, using read.delim and bind_rows.
data.table
The function rbindlist function from the package data.table also allows to merge the datasets contained in a list. In combination with fread (from the same package), it can be very fast.
What method is the fastest?
I will compare how long each combination of importing, vectorising, and merging functions needs to import 50 data sets with 10 columns and 10,000 rows each. Additionally, I will compare the performance of each method when working with CSV (.csv) and TSV (.txt) files. For each method, I will repeat the process 100 times, measuring how long it takes from the moment we list the extant files in the folder to the moment we finish merging the data sets. Here are the results:

For more detail:
| Time taken to import and merge | |||||||
|---|---|---|---|---|---|---|---|
| 50 datasets with 10 columns and 10,000 rows each | |||||||
| package | for loop | lapply | purrr::map | ||||
| M | SD | M | SD | M | SD | ||
| do.call - .csv | |||||||
| base | 1.34 | 0.06 | 1.11 | 0.07 | 0.15 | 0.03 | |
| data.table | 1.33 | 0.39 | 0.88 | 0.04 | 0.17 | 0.02 | |
| readr | 1.38 | 0.05 | 1.07 | 0.13 | 0.15 | 0.02 | |
| mean | — | 1.35 | 0.17 | 1.02 | 0.08 | 0.15 | 0.02 |
| dplyr::bind_rows - .csv | |||||||
| base | 1.35 | 0.16 | 0.99 | 0.14 | 0.15 | 0.01 | |
| data.table | 1.14 | 0.04 | 0.84 | 0.06 | 0.16 | 0.01 | |
| readr | 1.25 | 0.14 | 0.91 | 0.04 | 0.14 | 0.00 | |
| mean | — | 1.25 | 0.11 | 0.91 | 0.08 | 0.15 | 0.01 |
| do.call - .txt | |||||||
| base | 1.18 | 0.04 | 0.88 | 0.04 | 0.17 | 0.02 | |
| data.table | 1.17 | 0.04 | 0.80 | 0.05 | 0.15 | 0.02 | |
| readr | 1.18 | 0.05 | 0.80 | 0.05 | 0.15 | 0.02 | |
| mean | — | 1.18 | 0.05 | 0.82 | 0.05 | 0.15 | 0.02 |
| dplyr::bind_rows - .txt | |||||||
| base | 1.13 | 0.03 | 0.84 | 0.06 | 0.20 | 0.02 | |
| data.table | 1.19 | 0.13 | 0.77 | 0.03 | 0.14 | 0.01 | |
| readr | 1.13 | 0.04 | 0.77 | 0.03 | 0.14 | 0.01 | |
| mean | — | 1.15 | 0.07 | 0.79 | 0.04 | 0.16 | 0.01 |
Figure 1 and Table Table 1 show the detailed timings The grand mean average time taken by all methods is ~2.12 seconds, but there are some differences.
- It doesn’t really matter what function we use to merge data sets: both
do.callanddplyr::bind_rowsperform roughly similarly. - What makes the biggest difference is what function we use to vectorise the importing operation across file names to import them.
purrr::mapis the fastest. Incredibly, is takes less than 0.3 seconds in all conditions. It is also the least sensitive to the format of the files and the function we use to import them. - The next vectorising function in terms of temporal efficiency is
lapply, which takes ~1.5 seconds. It performs slightly better when working with .txt files, in that when working with .csv files its performance depends on what method we use to import them:data.table::freadis much faster than its base andreadrcompetitors. This post by Daniele Cook sheds some light into the advantage ofdata.tableover other importing functions, also covering thevroompackage, which this post doesn’t cover. - Using for loops looks like the least efficient method for iterating across data sets when importing data. It also shows a similar profile than
lapply:data.table::freadperforms a bit better than the rest.
Conclusion
Under the scenario under which I have simulated the data, it seems that using purrr::map in combination with do.call or dplyr::bind_rows to merge data sets is the most efficient method in terms of time. When using said combination, it doesn’t matter what function we use to import files, but data.table::fread seems like the best choice, as it is also the most flexible (take a look at the documentation of data.table to see all the features it offers).
If I have time, I may add another two dimensions: number of rows in the files and number of files, although I dare say similar results are to be expected. If anything, I would say that differences may become greater as file size and number of files increase. Also, it would be interesting to test if pre-allocating the elements of the vector in the for loop speeds up the process (see here what I mean). We shall see.
Hope this was useful, if not interesting!
Code
Session info
R version 4.3.3 (2024-02-29 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 11 x64 (build 22631)
Matrix products: default
locale:
[1] LC_COLLATE=English_United Kingdom.utf8
[2] LC_CTYPE=English_United Kingdom.utf8
[3] LC_MONETARY=English_United Kingdom.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United Kingdom.utf8
time zone: Europe/Berlin
tzcode source: internal
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] gt_0.10.1 ggsci_3.2.0 lubridate_1.9.3 forcats_1.0.0
[5] stringr_1.5.1 readr_2.1.5 tidyr_1.3.1 tibble_3.2.1
[9] ggplot2_3.5.1 tidyverse_2.0.0 data.table_1.15.4 purrr_1.0.2
[13] dplyr_1.1.4
loaded via a namespace (and not attached):
[1] gtable_0.3.5 jsonlite_1.8.8 compiler_4.3.3 renv_1.0.5
[5] tidyselect_1.2.1 xml2_1.3.6 scales_1.3.0 yaml_2.3.9
[9] fastmap_1.2.0 R6_2.5.1 commonmark_1.9.1 labeling_0.4.3
[13] generics_0.1.3 knitr_1.48 htmlwidgets_1.6.4 munsell_0.5.1
[17] pillar_1.9.0 tzdb_0.4.0 rlang_1.1.4 utf8_1.2.4
[21] stringi_1.8.4 xfun_0.46 sass_0.4.9 timechange_0.3.0
[25] cli_3.6.3 withr_3.0.0 magrittr_2.0.3 digest_0.6.36
[29] grid_4.3.3 markdown_1.13 hms_1.1.3 lifecycle_1.0.4
[33] vctrs_0.6.5 evaluate_0.24.0 glue_1.7.0 farver_2.1.2
[37] fansi_1.0.6 colorspace_2.1-0 rmarkdown_2.27 tools_4.3.3
[41] pkgconfig_2.0.3 htmltools_0.5.8.1Reuse
Citation
@online{garcia-castro2020,
author = {Garcia-Castro, Gonzalo},
title = {Importing Data from Multiple Files Simultaneously in {R}},
date = {2020-07-05},
url = {http://github.com/gongcastro/gongcastro.github.io/blog/importing-data-multiple/importing-data-multiple.html},
langid = {en}
}